
Discover in this article how we can quantify nearly 74 critical proteins with the combination of LC-MS and Scout MRM.
Introduction & strategy
Nowadays, cells are widely used to develop various therapeutics. But, during the Downstream process (DSP), Host Cell Proteins (HCPs) could be co-purified with the biotherapeutic. Copurification, in a chemical or biochemical context, is the physical separation by chromatography or other purification technique of two or more substances of interest from other contaminating substances. Thus, some known HCPs can affect drug safety or efficiency either because of:
- their high immunogenic power;
- their ability to degrade the final product (drug or adjuvant).
Consequently, it is mandatory to control the presence of such HCPs in drug substance (DS), or after the last DSP steps.
Liquid Chromatography–Mass Spectrometry (LC-MS) has become the complementary method of the ELISA method for Host Cell Protein quantification in biotherapeutics. Batch-to-batch comparison or DSP evaluation through Host Cell Protein analysis are more and more obtained using this analytical method.
Broad approaches using Data dependent or Data independent MS acquisition modes (DDA, DIA) correspond to classical methods to obtain great information such as HCPs identities. But these high throughput results require to be associated with a validation method to deliver expected and robust outcomes. Targeted Mass Spectrometry analysis, using selected/multiple reaction monitoring (SRM/MRM) or parallel reaction monitoring (PRM), offers the capability to measure and quantify a finite set of proteins with
- high sensitivity;
- specificity;
- reproducibility.
This is why the combination of the two techniques is interesting to obtain more reliable results on a large number of proteins. Information regarding the presence of critical HCPs in drug substances, but also at different steps of the DSP, may support the drafting and validation of regulatory records.
In this study, we developed a powerful solution using the READYBEADS technology associated with the SCOUT-MRM method to screen 74 critical HCPs in different matrices. A DS made up of elements produced in the Community Health Officer (CHO) cell line, was screened using the developed HCP-targeted SCOUT-MRM method. The DS is made up of:
- a therapeutic antibody;
- a post protein A intermediate step;
- a cell culture harvest from the same therapeutic antibody production.
Monitor 74 critical HCPs: our method
To conduct our experiment, 74 critical HCPs were extracted from an in-house HCP database and other knowledge obtained from the literature, including:
- The immunogenic PLBL2 lipase: has an impact on immunogenicity.
-
- The HTRA1 protease: high-temperature requirement protein A1, so named because of its essential role in thermal tolerance in Escherichia coli.
- The BIP proteins: binding immunoglobulin proteins, chaperones of newly synthesised proteins.
- 71Kda Heat shock cognate protein: acts as a repressor of transcriptional activation.
82 peptides were screened by SCOUT-SRM approach and the 82 corresponding heavy internal standard peptides were adsorbed on READYBEADS, according to a well-defined and patented process (patented by ANAQUANT) (Figure 1). Since READYBEADS technology offers valuable reproducibility, the normalized HCP signals could be compared between different samples.
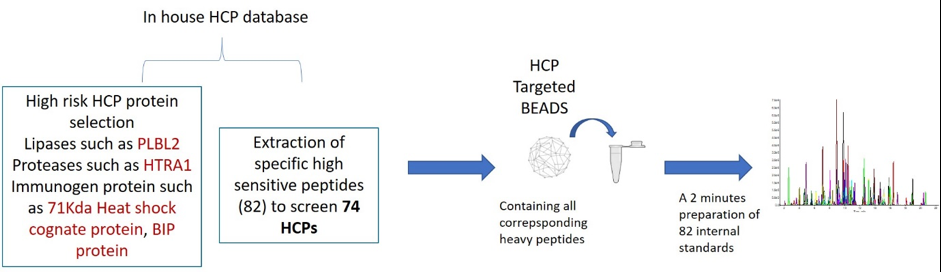
Figure 1: HCP Targeted method implementation
Results of the analytical experiment
Analytical strategy was performed on:
- the therapeutic mAb harvest sample;
- its post-protein A purified version;
- the DS sample obtained after the whole purification process.
The ultimate objective was to screen all 74 well-known HCPs and to validate the best downstream process to use after post-protein A purification.
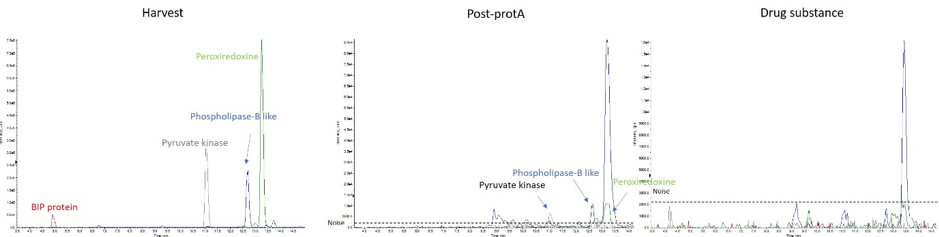
Figure 2: Extracted chromatogram of PBPL2 (blue), Pyruvate kinase (grey), Protein chaperon BIP (Red) in harvest mAb sample, post prot A mAb sample, and DS mAb sample.
Among all 74 well-described HCPs screened, 50 were identified in the harvest sample, 15 remain present after protein A purification step, while no risky HCPs were identified in the corresponding DS sample (see table below).
| Risky/well-known HCPs screened | HCPs identified in Harvest | HCPs identified in post protein A | HCPs identified in DS1 |
| 74 | 50 | 15 | 0 |
Concluding Remark
Thanks to our developed method, we offer the possibility to screen 74 critical HCPs in any samples using a simple robust method that could be easily and routinely implemented in any laboratory.
SCOUT-MRM allows achieving a robust highly multiplexed MRM assay. The entire SCOUT-MRM solution using HCP Targeted BEADS associated with SCOUT READYBEADS® allows validating specifically the critical HCPs identification and comparison between different samples and matrices.
The LC-MS technique is not limited to these 72 proteins. It could be used for the specific dosage and quantification of 400 to 500 proteins.
Thanks to this approach, the analytical method does not need to be developed specifically for each matrix but could be easily adapted, whatever the therapeutic drug or the purification step to screen. In addition, this method could also be easily implemented routinely in your laboratory.
The outcome: simplify your processes and save time!
 Anaquant HCP analysis I Protein characterisation I Protein analysis
Anaquant HCP analysis I Protein characterisation I Protein analysis